Providing Data for Plots and Tables¶
No data visualization is possible without the underlying data to be represented.
In this section, the various ways of providing data for plots is explained, from
passing data values directly to creating a ColumnDataSource and filtering using
a CDSView.
Providing data directly¶
In Bokeh, it is possible to pass lists of values directly into plotting functions.
In the example below, the data, x_values and y_values, are passed directly
to the circle plotting method (see Plotting with Basic Glyphs for more examples).
from bokeh.plotting import figure
x_values = [1, 2, 3, 4, 5]
y_values = [6, 7, 2, 3, 6]
p = figure()
p.circle(x=x_values, y=y_values)
When you pass in data like this, Bokeh works behind the scenes to make a
ColumnDataSource for you. But learning to create and use the ColumnDataSource
will enable you access more advanced capabilites, such as streaming data,
sharing data between plots, and filtering data.
ColumnDataSource¶
The ColumnDataSource is the core of most Bokeh plots, providing the data
that is visualized by the glyphs of the plot. With the ColumnDataSource,
it is easy to share data between multiple plots and widgets, such as the
DataTable. When the same ColumnDataSource is used to drive multiple
renderers, selections of the data source are also shared. Thus it is possible
to use a select tool to choose data points from one plot and have them automatically
highlighted in a second plot (Linked selection).
At the most basic level, a ColumnDataSource is simply a mapping between column
names and lists of data. The ColumnDataSource takes a data parameter which is a dict,
with string column names as keys and lists (or arrays) of data values as values. If one positional
argument is passed in to the ColumnDataSource initializer, it will be taken as data. Once the
ColumnDataSource has been created, it can be passed into the source parameter of
plotting methods which allows you to pass a column’s name as a stand in for the data values:
from bokeh.plotting import figure
from bokeh.models import ColumnDataSource
data = {'x_values': [1, 2, 3, 4, 5],
'y_values': [6, 7, 2, 3, 6]}
source = ColumnDataSource(data=data)
p = figure()
p.circle(x='x_values', y='y_values', source=source)
Note
There is an implicit assumption that all the columns in a given ColumnDataSource
all have the same length at all times. For this reason, it is usually preferable to
update the .data property of a data source “all at once”.
Pandas¶
The data parameter can also be a Pandas DataFrame or GroupBy object.
source = ColumnDataSource(df)
If a DataFrame is used, the CDS will have columns corresponding to the columns of
the DataFrame. The index of the DataFrame will be reset, so if the DataFrame
has a named index column, then CDS will also have a column with this name. However,
if the index name is None, then the CDS will be assigned a generic name.
It will be index if it is available, and level_0 otherwise.
Pandas MultiIndex¶
All MultiIndex columns and indices will be flattened before forming the
ColumnsDataSource. For the index, an index of tuples will be created, and the
names of the MultiIndex joined with an underscore. The column names will also
be joined with an underscore. For example:
df = pd.DataFrame({('a', 'b'): {('A', 'B'): 1, ('A', 'C'): 2},
('b', 'a'): {('A', 'C'): 7, ('A', 'B'): 8},
('b', 'b'): {('A', 'D'): 9, ('A', 'B'): 10}})
cds = ColumnDataSource(df)
will result in a column named index with [(A, B), (A, C), (A, D)] and columns
named a_b, b_a, and b_b. This process will fail for non-string column names,
so flatten the DataFrame manually in that case.
Pandas GroupBy¶
group = df.groupby(('colA', 'ColB'))
source = ColumnDataSource(group)
If a GroupBy object is used, the CDS will have columns corresponding to the result of
calling group.describe(). The describe method generates columns for statistical measures
such as mean and count for all the non-grouped orginal columns. The resulting DataFrame
has MultiIndex columns with the original column name and the columputed measure, so it
will be flattened using the aforementioned scheme. For example, if a
DataFrame has columns 'year' and 'mpg'. Then passing df.groupby('year')
to a CDS will result in columns such as 'mpg_mean'
Note this capability to adapt GroupBy objects may only work with Pandas >=0.20.0.
Streaming¶
ColumnDataSource streaming is an efficient way to append new data to a CDS. By using the
stream method, Bokeh only sends new data to the browser instead of the entire dataset.
The stream method takes a new_data parameter containing a dict mapping column names
to sequences of data to be appended to the respective columns. It additionally takes an optional
argument rollover, which is the maximum length of data to keep (data from the beginning of the
column will be discarded). The default rollover value of None allows data to grow unbounded.
source = ColumnDataSource(data=dict(foo=[], bar=[]))
# has new, identical-length updates for all columns in source
new_data = {
'foo' : [10, 20],
'bar' : [100, 200],
}
source.stream(new_data)
For an example that uses streaming, see examples/app/ohlc.
Patching¶
ColumnDataSource patching is an efficient way to update slices of a data source. By using the
patch method, Bokeh only needs to send new data to the browser instead of the entire dataset.
The patch method should be passed a dict mapping column names to list of tuples that represent
a patch change to apply.
The tuples that describe patch changes are of the form:
(index, new_value) # replace a single column value
# or
(slice, new_values) # replace several column values
For a full example, see examples/howto/patch_app.py.
Transforming Data¶
We have seen above how data can be added to a ColumnDataSource to drive
Bokeh plots. This can include raw data or data that we explicitly transform
ourselves, for example a column of colors created to control how the Markers
in a scatter plot should be shaded. It is also possible to specify transforms
that only occur in the browser. This can be useful to reduce both code (i.e.
not having to color map data by hand) as well as the amount of data that has to
be sent into the browser (only the raw data is sent, and colormapping occurs
in the client).
In this section we examine some of the different transform objects that are available.
Colors¶
To perform linear colormapping in the browser, the
linear_cmap() function may be used. It accepts the name
of a ColumnDataSource column to colormap, a palette (which can be a built-in
palette name, or an actual list of colors), and min/max values for the color
mapping range. The result can be passed to a color property on glyphs:
fill_color=linear_cmap('counts', 'Viridis256', min=0, max=10)
A complete example is shown here:
import numpy as np
from bokeh.plotting import figure, show
from bokeh.transform import linear_cmap
from bokeh.util.hex import hexbin
n = 50000
x = np.random.standard_normal(n)
y = np.random.standard_normal(n)
bins = hexbin(x, y, 0.1)
p = figure(tools="", match_aspect=True, background_fill_color='#440154')
p.grid.visible = False
p.hex_tile(q="q", r="r", size=0.1, line_color=None, source=bins,
fill_color=linear_cmap('counts', 'Viridis256', 0, max(bins.counts)))
show(p)
Besides linear_cmap() there is also
log_cmap() to perform color mapping on a log scale, as
well as factor_cmap() to colormap categorical data (see
the example below).
Markers¶
It is also possible to map categorical data to marker types. The example
below shows the use of factor_mark() to display different
markers or different categories in the input data. It also demonstrates the use
of factor_cmap() to colormap those same categories:
from bokeh.plotting import figure, show
from bokeh.sampledata.iris import flowers
from bokeh.transform import factor_cmap, factor_mark
SPECIES = ['setosa', 'versicolor', 'virginica']
MARKERS = ['hex', 'circle_x', 'triangle']
p = figure(title = "Iris Morphology")
p.xaxis.axis_label = 'Petal Length'
p.yaxis.axis_label = 'Sepal Width'
p.scatter("petal_length", "sepal_width", source=flowers, legend="species", fill_alpha=0.4, size=12,
marker=factor_mark('species', MARKERS, SPECIES),
color=factor_cmap('species', 'Category10_3', SPECIES))
show(p)
Note
The factor_mark() transform is primarily only useful
with the scatter glyph method, since only the Scatter glyph can be
parameterized by marker type.
CustomJSTransform¶
In addition to built-in transforms above, there is also a CustomJSTransform
that allows for specifying arbitary JavaScript code to perform a tranform step
on ColumnDataSource data. Typically, the v_func (for “vectorized” function)
is provided. (Less commonly a scalar equivalent func may also be needed).
The v_func code should expect an array of inputs in the variable xs, and
return a JavaScript array with the transformed values:
v_func = """
const first = xs[0]
const norm = new Float64Array(xs.length)
for (let i = 0; i < xs.length; i++) {
norm[i] = xs[i] / first
}
return norm
"""
normalize = CustomJSTransform(v_func=v_func)
plot.line(x='aapl_date', y=transform('aapl_close', normalize), line_width=2,
color='#cf3c4d', alpha=0.6,legend="Apple", source=aapl_source)
The above code converts raw price data into a sequence of normalized returns relative to the first data point. The full result is shown below:
Filtering Data¶
It’s often desirable to focus in on a portion of data that has been subsampled or filtered from a larger dataset. Bokeh allows you to specify a view of a data source that represents a subset of data. By having a view of the data source, the underlying data doesn’t need to be changed and can be shared across plots. The view consists of one or more filters that select the rows of the data source that should be bound to a specific glyph.
To plot with a subset of data, you can create a CDSView and pass it in as a view
argument to the renderer-adding methods on the Figure, such as figure.circle. The
CDSView has two properties, source and filters. source is the ColumnDataSource
that the view is associated with. filters is a list of Filter objects, listed and
described below.
from bokeh.plotting import figure
from bokeh.models import ColumnDataSource, CDSView
source = ColumnDataSource(some_data)
view = CDSView(source=source, filters=[filter1, filter2])
p = figure()
p.circle(x="x", y="y", source=source, view=view)
IndexFilter¶
The IndexFilter is the simplest filter type. It has an indices property which is a
list of integers that are the indices of the data you want to be included in the plot.
from bokeh.layouts import gridplot
from bokeh.models import ColumnDataSource, CDSView, IndexFilter
from bokeh.plotting import figure, show
source = ColumnDataSource(data=dict(x=[1, 2, 3, 4, 5], y=[1, 2, 3, 4, 5]))
view = CDSView(source=source, filters=[IndexFilter([0, 2, 4])])
tools = ["box_select", "hover", "reset"]
p = figure(plot_height=300, plot_width=300, tools=tools)
p.circle(x="x", y="y", size=10, hover_color="red", source=source)
p_filtered = figure(plot_height=300, plot_width=300, tools=tools)
p_filtered.circle(x="x", y="y", size=10, hover_color="red", source=source, view=view)
show(gridplot([[p, p_filtered]]))
BooleanFilter¶
A BooleanFilter selects rows from a data source through a list of True or False values
in its booleans property.
from bokeh.layouts import gridplot
from bokeh.models import ColumnDataSource, CDSView, BooleanFilter
from bokeh.plotting import figure, show
source = ColumnDataSource(data=dict(x=[1, 2, 3, 4, 5], y=[1, 2, 3, 4, 5]))
booleans = [True if y_val > 2 else False for y_val in source.data['y']]
view = CDSView(source=source, filters=[BooleanFilter(booleans)])
tools = ["box_select", "hover", "reset"]
p = figure(plot_height=300, plot_width=300, tools=tools)
p.circle(x="x", y="y", size=10, hover_color="red", source=source)
p_filtered = figure(plot_height=300, plot_width=300, tools=tools,
x_range=p.x_range, y_range=p.y_range)
p_filtered.circle(x="x", y="y", size=10, hover_color="red", source=source, view=view)
show(gridplot([[p, p_filtered]]))
GroupFilter¶
The GroupFilter allows you to select rows from a dataset that have a specific value for
a categorical variable. The GroupFilter has two properties, column_name, the name of
column in the ColumnDataSource, and group, the value of the column to select for.
In the example below, flowers contains a categorical variable species which is
either setosa, versicolor, or virginica.
from bokeh.layouts import gridplot
from bokeh.models import ColumnDataSource, CDSView, GroupFilter
from bokeh.plotting import figure, show
from bokeh.sampledata.iris import flowers
source = ColumnDataSource(flowers)
view1 = CDSView(source=source, filters=[GroupFilter(column_name='species', group='versicolor')])
plot_size_and_tools = {'plot_height': 300, 'plot_width': 300,
'tools':['box_select', 'reset', 'help']}
p1 = figure(title="Full data set", **plot_size_and_tools)
p1.circle(x='petal_length', y='petal_width', source=source, color='black')
p2 = figure(title="Setosa only", x_range=p1.x_range, y_range=p1.y_range, **plot_size_and_tools)
p2.circle(x='petal_length', y='petal_width', source=source, view=view1, color='red')
show(gridplot([[p1, p2]]))
CustomJSFilter¶
You can also create a CustomJSFilter with your own functionality. To do this, use JavaScript,
TypeScript or CoffeeScript to write code that returns either a list of indices or a list of
booleans that represents the filtered subset. The ColumnDataSource that is associated
with the CDSView this filter is added to will be available at render time with the
variable source.
Javascript¶
To create a CustomJSFilter with custom functionality written in JavaScript,
pass in the JavaScript code as a string to the parameter code:
custom_filter = CustomJSFilter(code='''
var indices = [];
// iterate through rows of data source and see if each satisfies some constraint
for (var i = 0; i < source.get_length(); i++){
if (source.data['some_column'][i] == 'some_value'){
indices.push(true);
} else {
indices.push(false);
}
}
return indices;
''')
Coffeescript¶
You can also write code for the CustomJSFilter in CoffeeScript, and
use the from_coffeescript class method, which accepts the code parameter:
custom_filter_coffee = CustomJSFilter.from_coffeescript(code='''
z = source.data['z']
indices = (i for i in [0...source.get_length()] when z[i] == 'b')
return indices
''')
AjaxDataSource¶
Bokeh server applications make it simple to update and stream data to data
sources, but sometimes it is desirable to have similar functionality in
standalone documents. The AjaxDataSource
provides this cabability.
The AjaxDataSource is configured with a URL to a REST endoint and a
polling interval. In the browser, the data source will request data from the
endpoint at the specified interval and update the data locally. Existing
data may either be replaced entirely, or appened to (up to a configurable
max_size). The endpoint that is supplied should return a JSON dict that
matches the standard ColumnDataSource format:
{
'x' : [1, 2, 3, ...],
'y' : [9, 3, 2, ...]
}
Otherwise, using an AjaxDataSource is identical to using a standard
ColumnDataSource:
source = AjaxDataSource(data_url='http://some.api.com/data',
polling_interval=100)
# Use just like a ColumnDataSource
p.circle('x', 'y', source=source)
A full example (shown below) can be seen at examples/howto/ajax_source.py
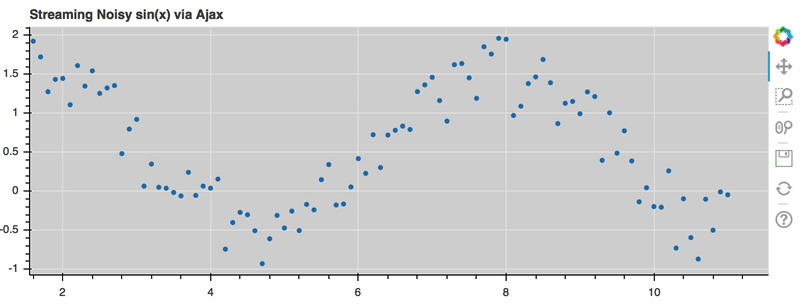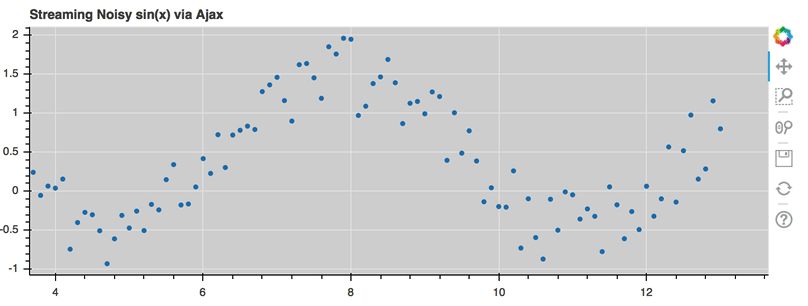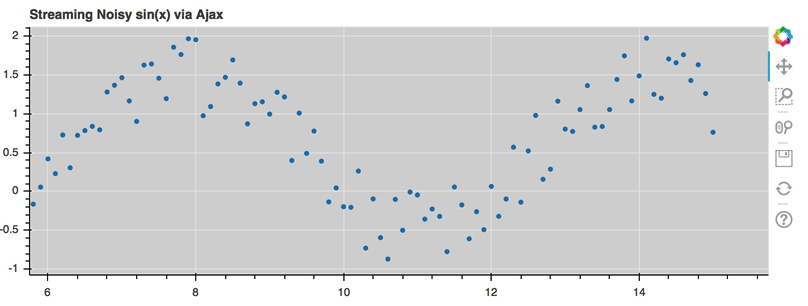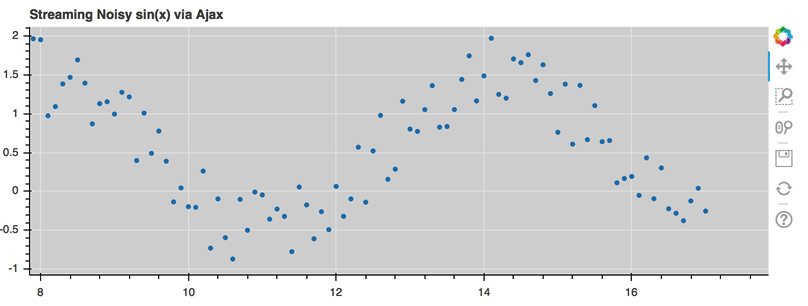
Linked selection¶
Using the same ColumnDataSource in the two plots below allows their selections to be
shared.
from bokeh.io import output_file, show
from bokeh.layouts import gridplot
from bokeh.models import ColumnDataSource
from bokeh.plotting import figure
output_file("brushing.html")
x = list(range(-20, 21))
y0 = [abs(xx) for xx in x]
y1 = [xx**2 for xx in x]
# create a column data source for the plots to share
source = ColumnDataSource(data=dict(x=x, y0=y0, y1=y1))
TOOLS = "box_select,lasso_select,help"
# create a new plot and add a renderer
left = figure(tools=TOOLS, plot_width=300, plot_height=300, title=None)
left.circle('x', 'y0', source=source)
# create another new plot and add a renderer
right = figure(tools=TOOLS, plot_width=300, plot_height=300, title=None)
right.circle('x', 'y1', source=source)
p = gridplot([[left, right]])
show(p)
Linked selection with filtered data¶
With the ability to specify a subset of data to be used for each glyph renderer, it is
easy to share data between plots even when the plots use different subsets of data.
By using the same ColumnDataSource, selections and hovered inspections of that data source
are automatically shared.
In the example below, a CDSView is created for the second plot that specifies the subset
of data in which the y values are either greater than 250 or less than 100. Selections in either
plot are automatically reflected in the other. And hovering on a point in one plot will highlight
the corresponding point in the other plot if it exists.
from bokeh.plotting import figure, output_file, show
from bokeh.layouts import gridplot
from bokeh.models import ColumnDataSource, CDSView, BooleanFilter
output_file("linked_selection_subsets.html")
x = list(range(-20, 21))
y0 = [abs(xx) for xx in x]
y1 = [xx**2 for xx in x]
# create a column data source for the plots to share
source = ColumnDataSource(data=dict(x=x, y0=y0, y1=y1))
# create a view of the source for one plot to use
view = CDSView(source=source, filters=[BooleanFilter([True if y > 250 or y < 100 else False for y in y1])])
TOOLS = "box_select,lasso_select,hover,help"
# create a new plot and add a renderer
left = figure(tools=TOOLS, plot_width=300, plot_height=300, title=None)
left.circle('x', 'y0', size=10, hover_color="firebrick", source=source)
# create another new plot, add a renderer that uses the view of the data source
right = figure(tools=TOOLS, plot_width=300, plot_height=300, title=None)
right.circle('x', 'y1', size=10, hover_color="firebrick", source=source, view=view)
p = gridplot([[left, right]])
show(p)
Other Data Types¶
Bokeh also has the capability to render network graph data and geographical data. For more information about how to set up the data for these types of plots, see Visualizing Network Graphs and Mapping Geo Data.
