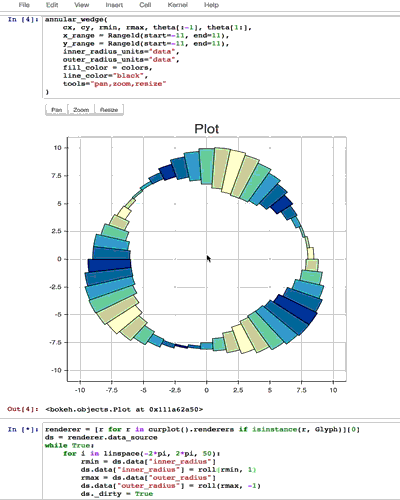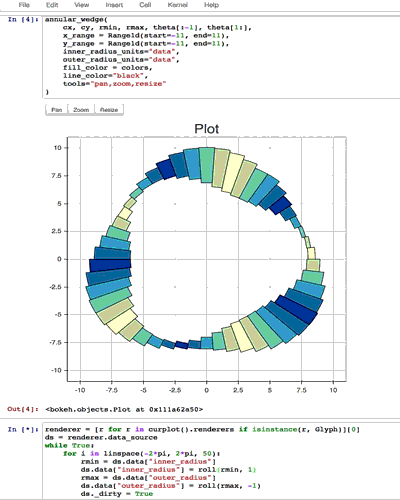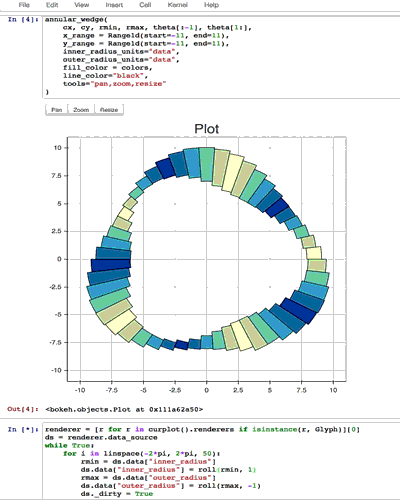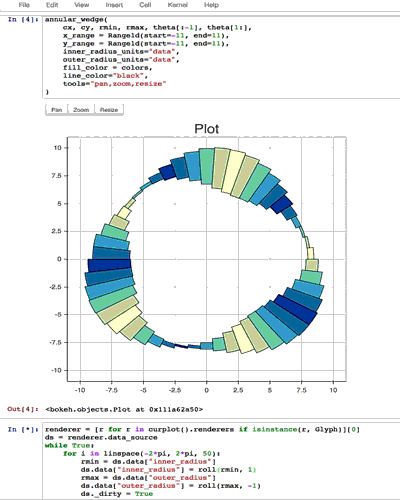
from collections import OrderedDict
from math import log, sqrt
import numpy as np
import pandas as pd
from six.moves import cStringIO as StringIO
from bokeh.plotting import *
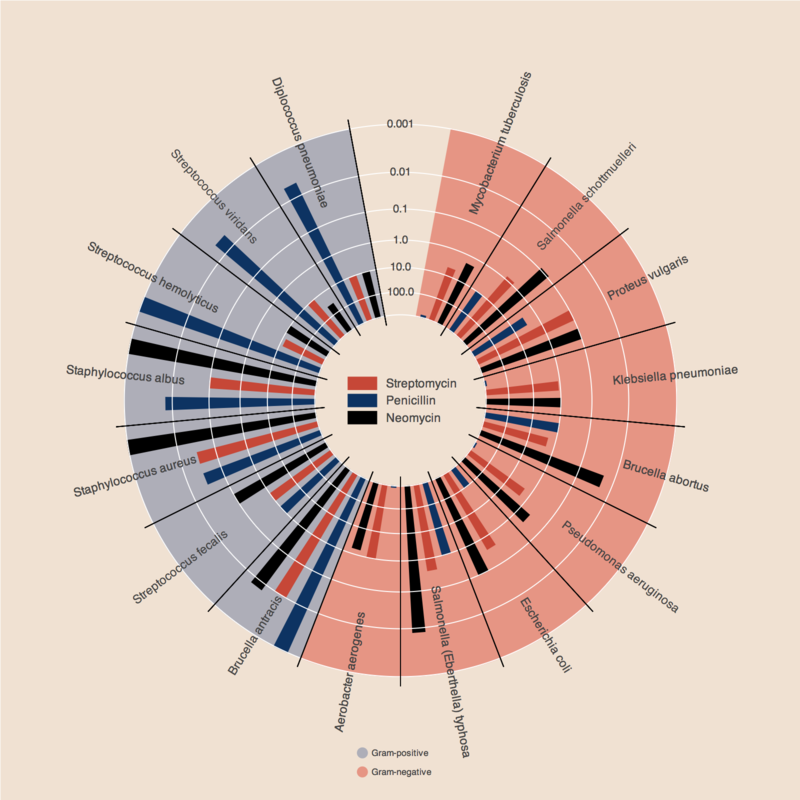
antibiotics = """
bacteria, penicillin, streptomycin, neomycin, gram
Mycobacterium tuberculosis, 800, 5, 2, negative
Salmonella schottmuelleri, 10, 0.8, 0.09, negative
Proteus vulgaris, 3, 0.1, 0.1, negative
Klebsiella pneumoniae, 850, 1.2, 1, negative
Brucella abortus, 1, 2, 0.02, negative
Pseudomonas aeruginosa, 850, 2, 0.4, negative
Escherichia coli, 100, 0.4, 0.1, negative
Salmonella (Eberthella) typhosa, 1, 0.4, 0.008, negative
Aerobacter aerogenes, 870, 1, 1.6, negative
Brucella antracis, 0.001, 0.01, 0.007, positive
Streptococcus fecalis, 1, 1, 0.1, positive
Staphylococcus aureus, 0.03, 0.03, 0.001, positive
Staphylococcus albus, 0.007, 0.1, 0.001, positive
Streptococcus hemolyticus, 0.001, 14, 10, positive
Streptococcus viridans, 0.005, 10, 40, positive
Diplococcus pneumoniae, 0.005, 11, 10, positive
"""
drug_color = OrderedDict([
("Penicillin", "#0d3362"),
("Streptomycin", "#c64737"),
("Neomycin", "black" ),
])
gram_color = {
"positive" : "#aeaeb8",
"negative" : "#e69584",
}
df = pd.read_csv(StringIO(antibiotics), skiprows=1, skipinitialspace=True)
width = 800
height = 800
inner_radius = 90
outer_radius = 300 - 10
minr = sqrt(log(.001 * 1E4))
maxr = sqrt(log(1000 * 1E4))
a = (outer_radius - inner_radius) / (minr - maxr)
b = inner_radius - a * maxr
def rad(mic):
return a * np.sqrt(np.log(mic * 1E4)) + b
big_angle = 2.0 * np.pi / (len(df) + 1)
small_angle = big_angle / 7
x = np.zeros(len(df))
y = np.zeros(len(df))